Request Samples
If you are logged in to the CSTN data portal, mark the samples of interest in the Search Grid and press the blue "Action" dropdown and choose "Request Samples" option.
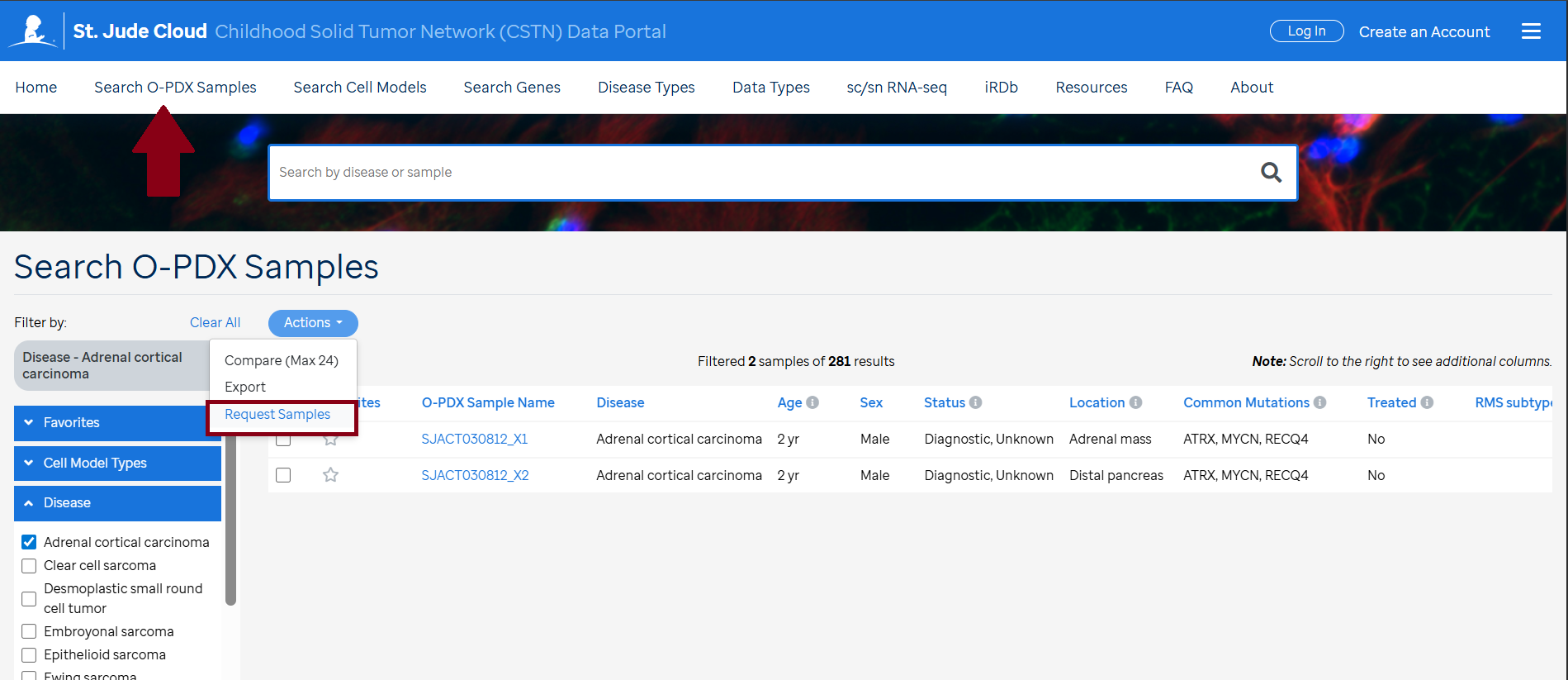
The request must be processed and approved by Michael Dyer, PhD; Dr. Alberto Pappo, MD; and the Childhood Solid Tumor Network (CSTN) committee. After approval, a Material Transfer Agreement (MTA) will be sent to your institution for signature and execution. If there is a need for further clarification prior to approval, we will contact you directly.
The typical process is as follows:
- Request is submitted;
- CSTN approves request;
- CSTN sends MTA to requesting institution;
- Requesting institution signs MTA, sends to CSTN for signature;
- CSTN confirms shipping details and ships samples;
- Request is closed.
Our goal is to share samples for timely use in order to drive more rapid discovery in the field, while minimizing the risk that valuable samples may be “stockpiled” for possible future use. We also prefer to keep numbers small to reduce the risk of loss or damage incurred by shipping large numbers of cryovials.
The timeline depends on your institution and on the time required to get the MTA signed and executed. Once the MTA is signed, we will contact you within a week and confirm shipment details.
The consent form under which the patient tumor samples were collected stipulates that all samples be de-identified and sent out under an MTA.
No. The CSTN actively tracks samples that have been distributed so that we may contact investigators should updated information about the sample become available. Please encourage any interested research colleagues to contact us directly by filling out a request form, and we will be happy to assist.
Stewart E*, Federico SM*, Chen X*, Shelat AA, Bradley C, Gordon B, Karlstrom A, Twarog NR, Clay MR, Bahrami A, Freeman BB 3rd, Xu B, Zhou X, Wu J, Honnell V, Ocarz M, Blankenship K, Dapper J, Mardis ER, Wilson RK, Downing J, Zhang J, Easton J, Pappo A, Dyer MA, Orthotopic patient-derived xenografts of paediatric solid tumours, Nature. 2017 Sep 7;549(7670):96-100
Enter the question in the comment section of the request form when you initially submit it, or send an email to CSTN@stjude.org.
No. We have been focused on propagating the xenografts and banking them down for sharing. In the future we may create cell lines from these samples.
You will receive the sample and an invoice with the official names and links to the vials. The associated SOPs for implantation, freezing and thawing is found on the CSTN Data portal under Resources.
You can subscribe to our newsletter to receive regular updates on new resources, data and advances emerging from the CSTN. We also share information about relevant clinical trials.
Commercial entities may contact CSTN@stjude.org for details.
Explore Samples
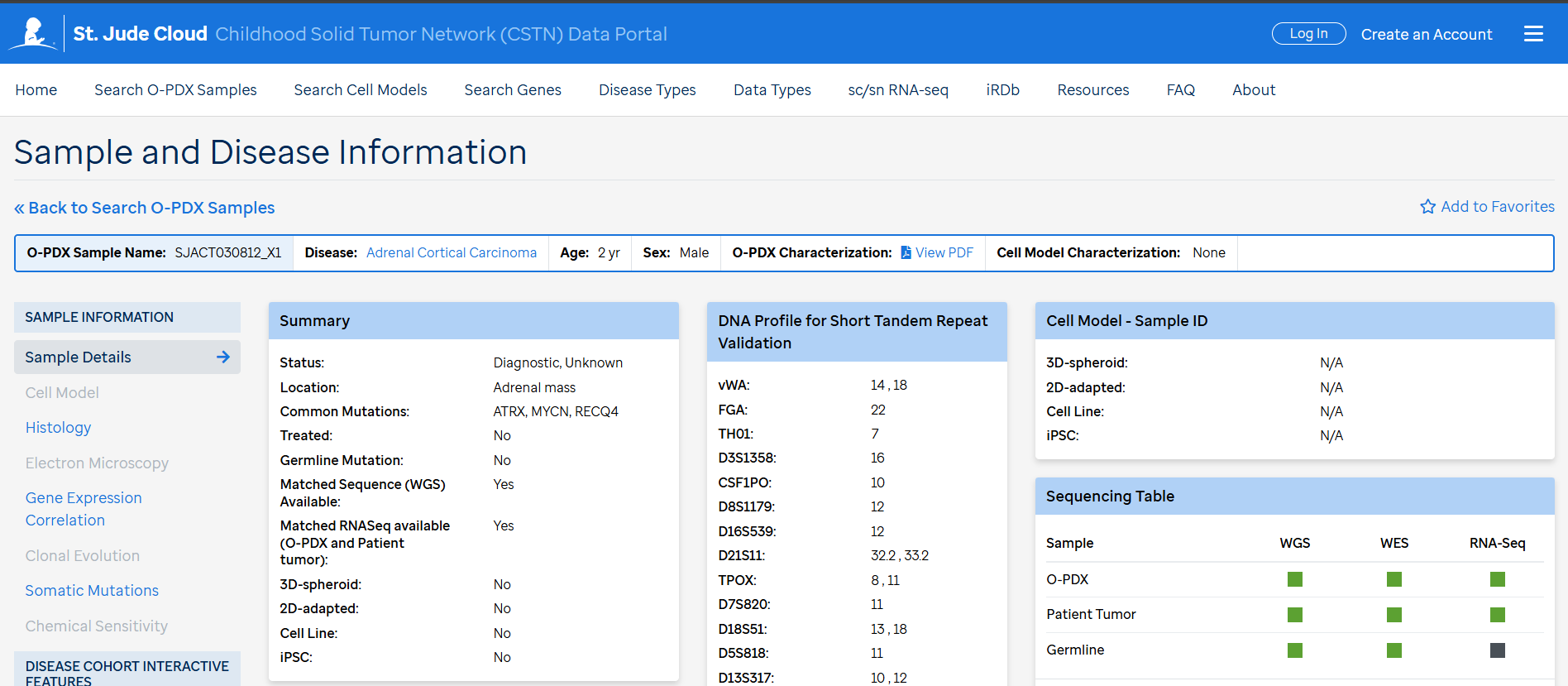
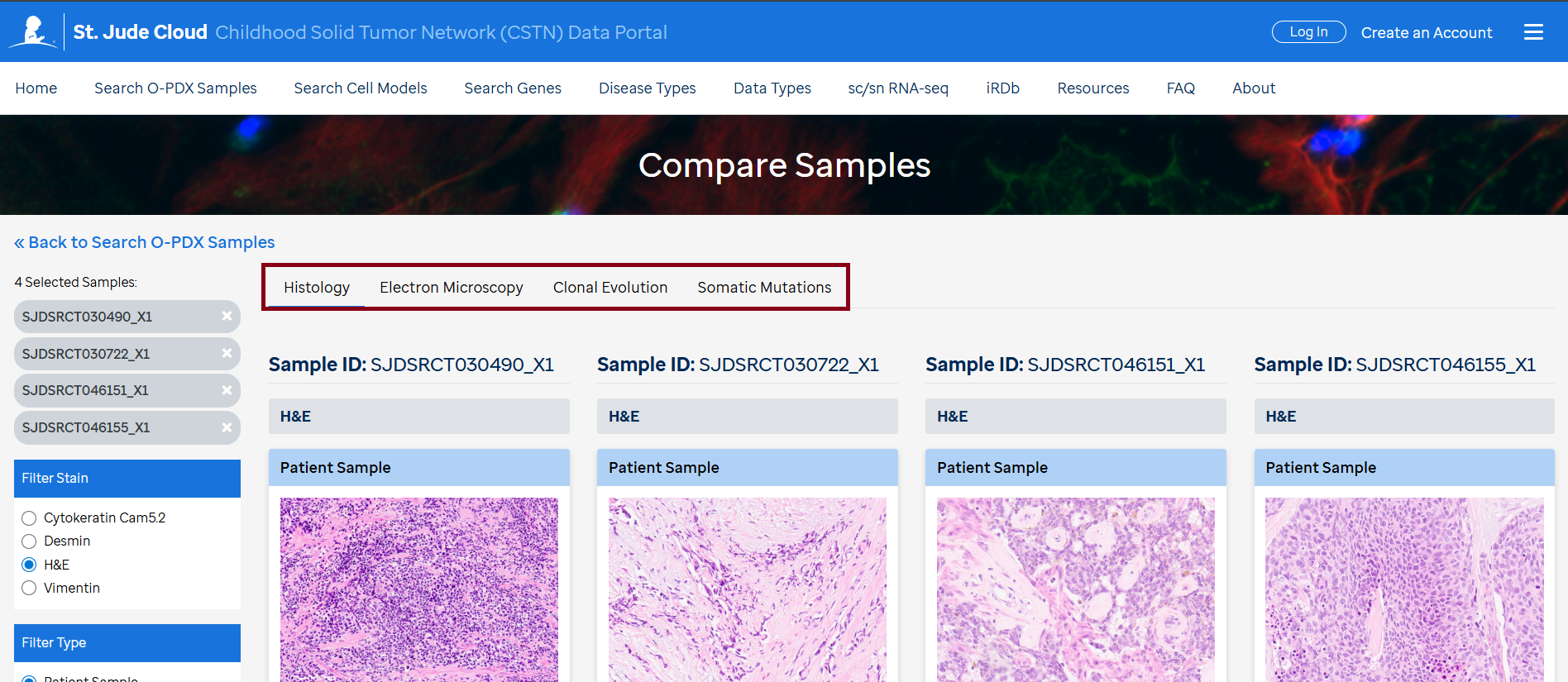
Characterization of the opdx samples include brief clinical information, STR DNA profile, histology (H&E, IHC), Electron Microscopy images, chemical sensitivity and clonal evolution. There is also comparison opdx-primary tumors as curated data in somatic mutations and gene expression correlation plots available under each sample.
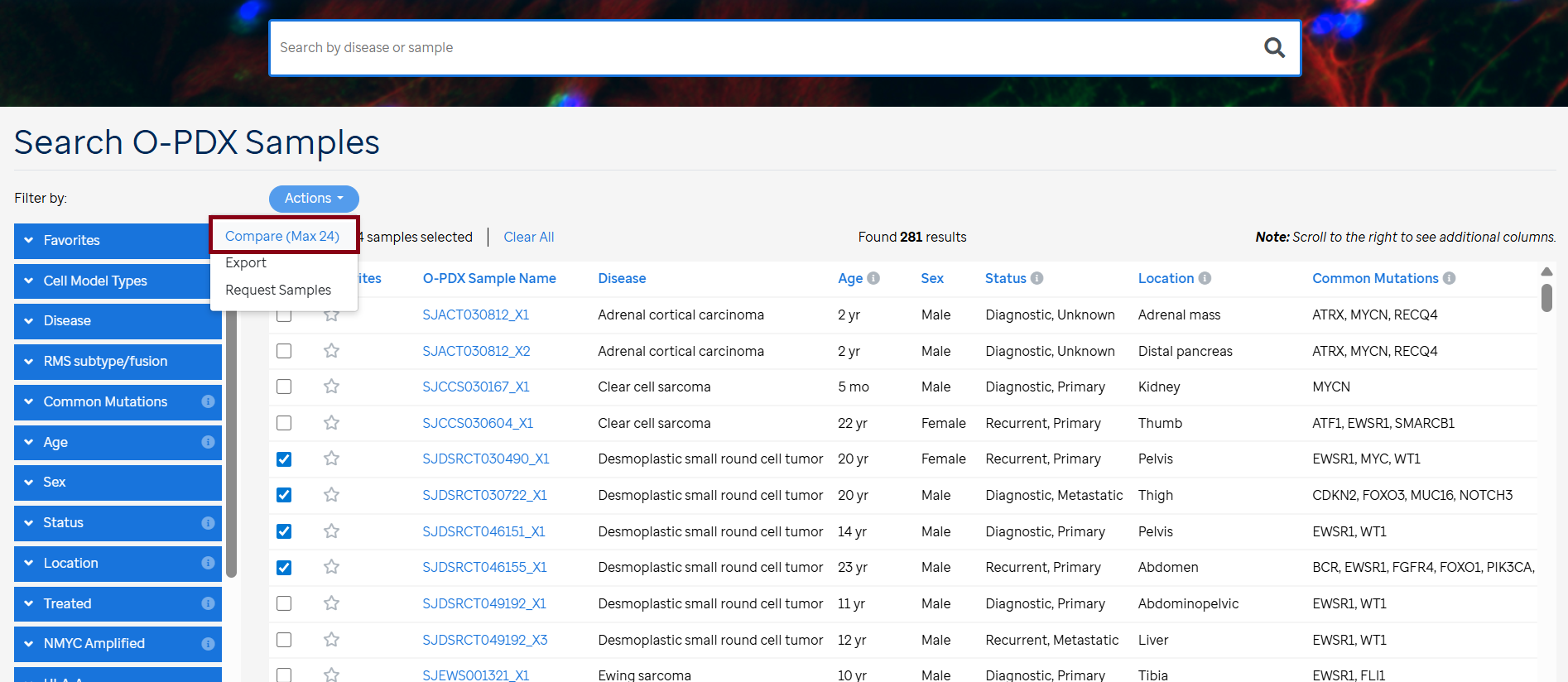
- Navigate to “Search O-PDX Samples”at the top of the page.
- Select the samples of interest by clicking the checkboxes in the leftmost column of the table.
- Click on the "Actions" dropdown above the table on the left and choose the "Compare (Max 24)" option.
- View the comparison of the selected samples side by side. Navigate between the Histology, Electron Microscopy, Clonal Evolution, and Somatic Mutations tabs for detailed comparisons.
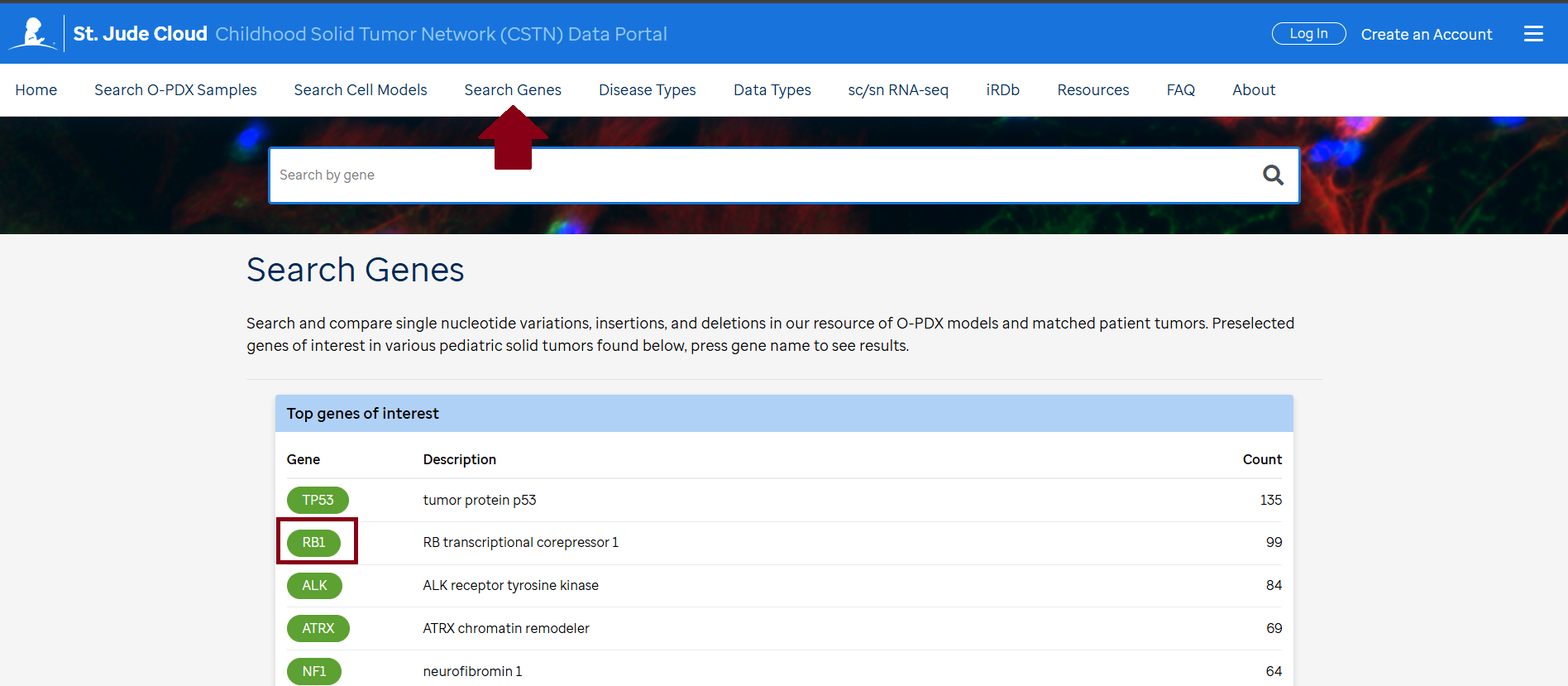
- Navigate to “Search Genes”at the top of the page.
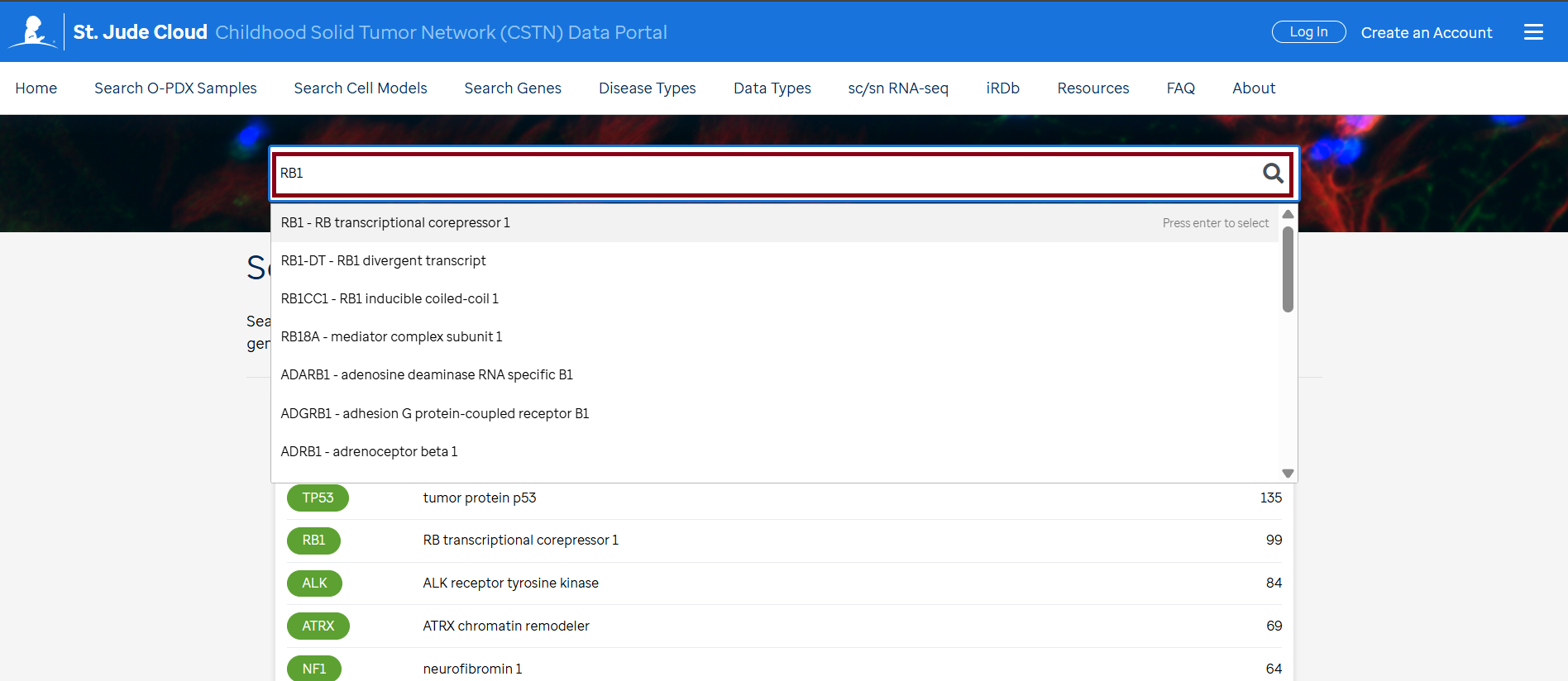
- Enter the gene(s) of interest in the "Search Box" on the page, then click the search icon.
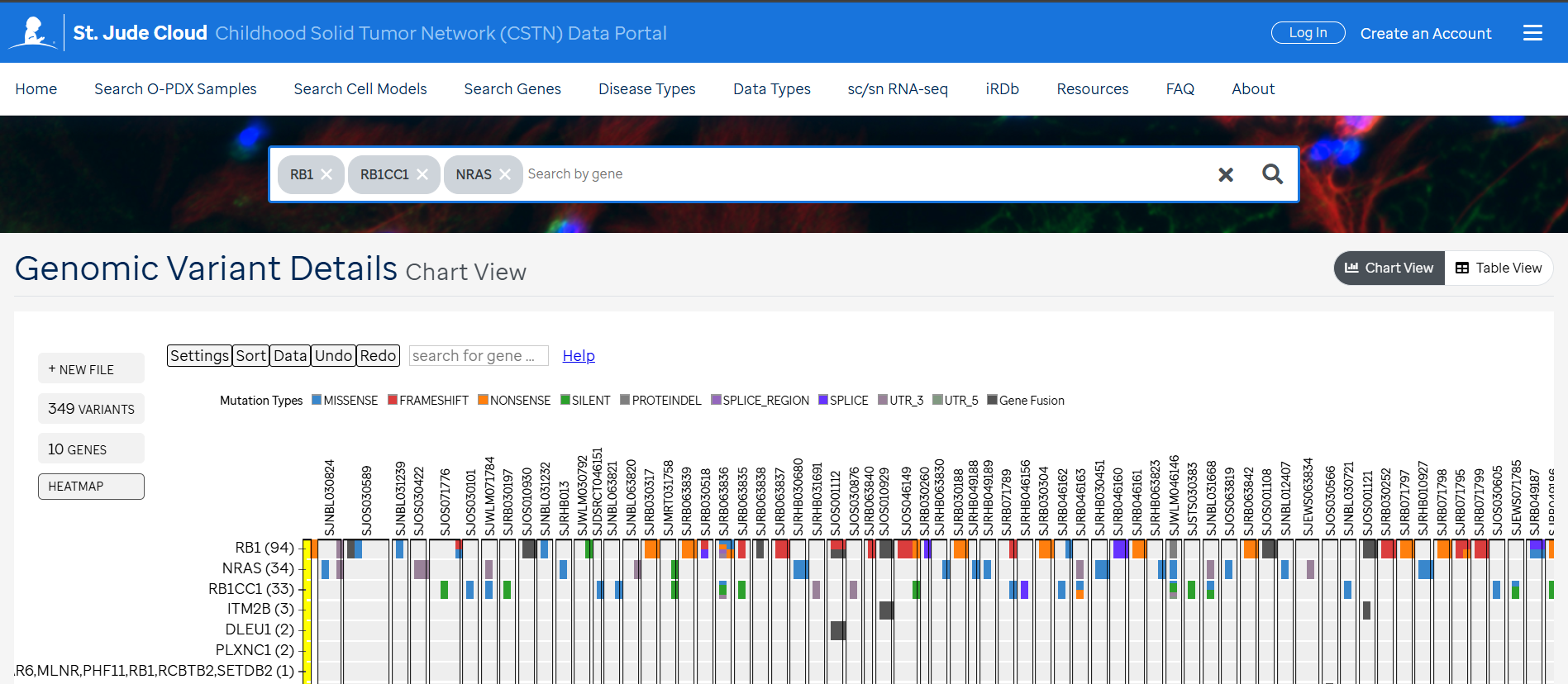
Access to curated data is available in the CSTN data portal under each individual sample (1) , curated disease mutation heatmaps in "Disease Cohort" under Data Types (2) and "Search Genes” (3).
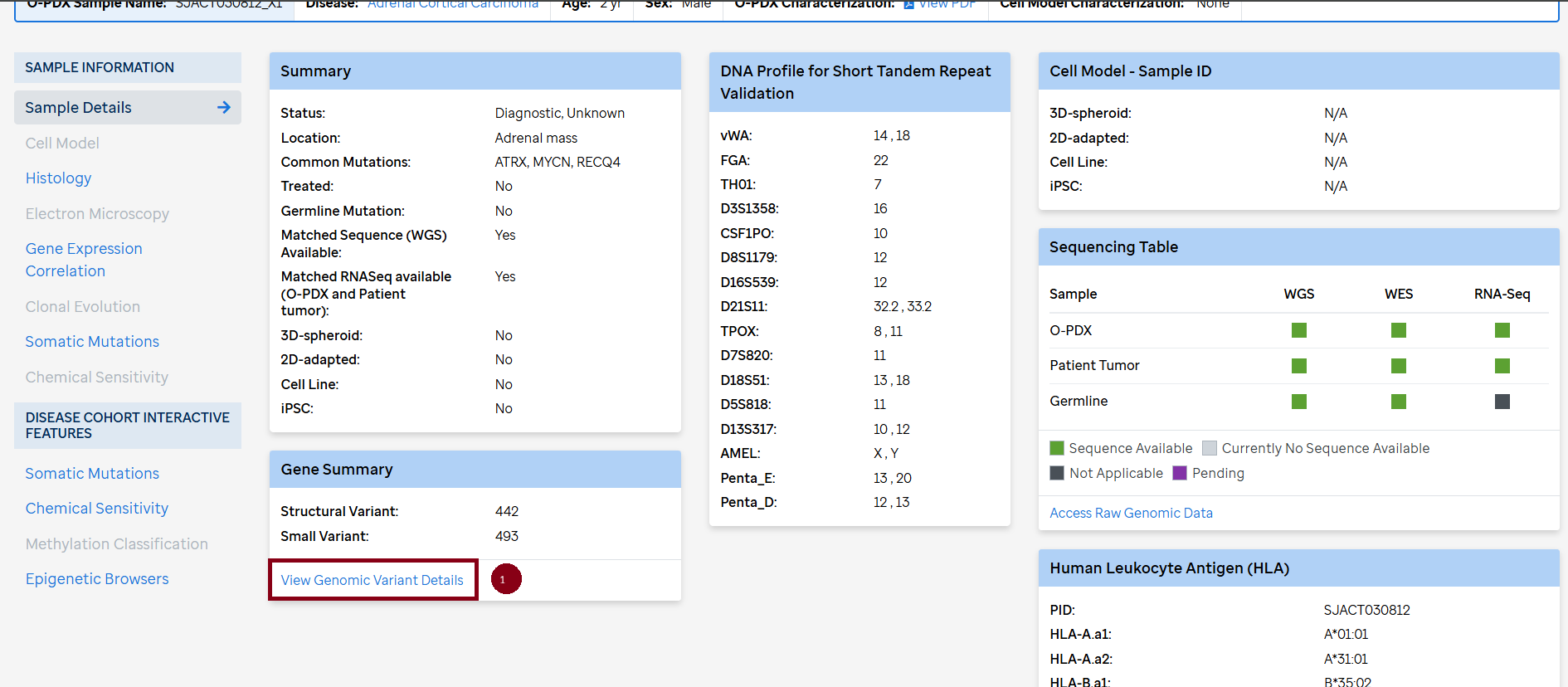
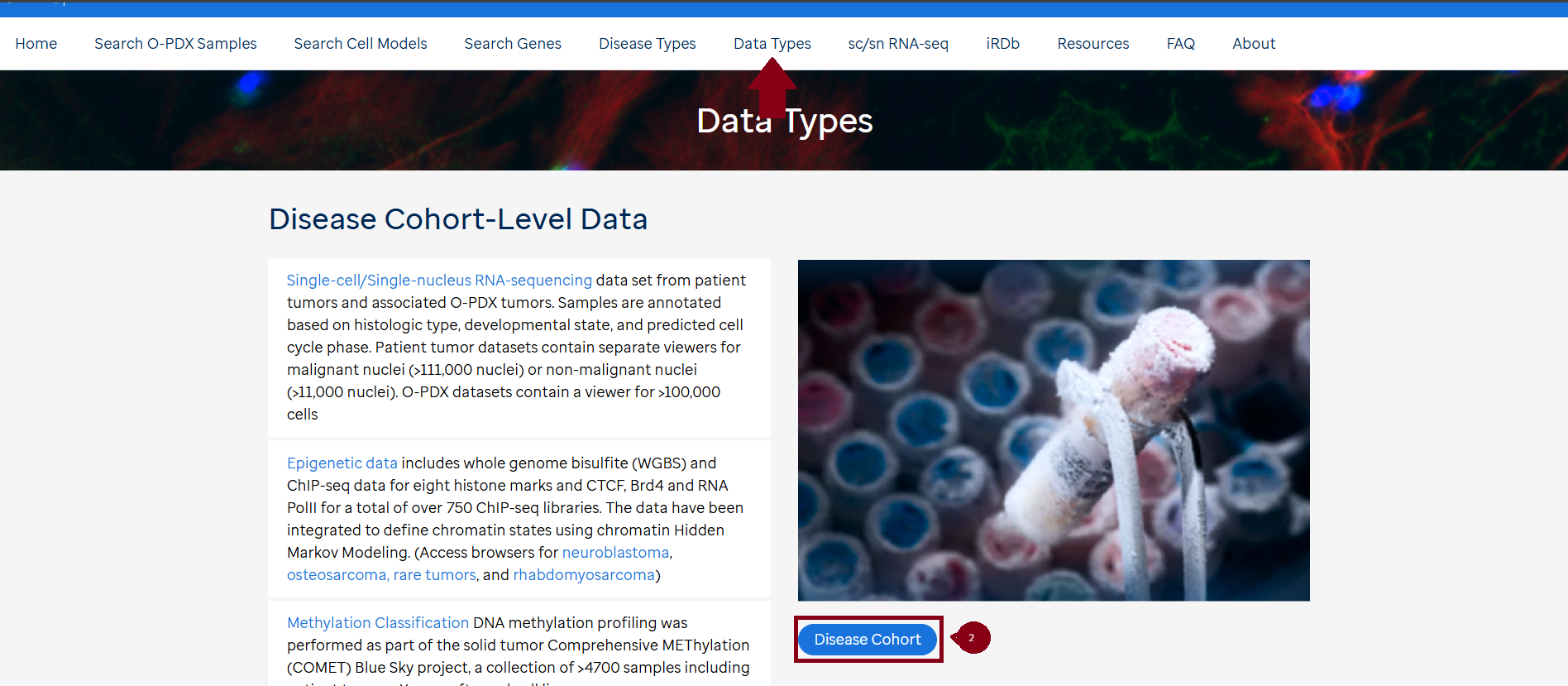
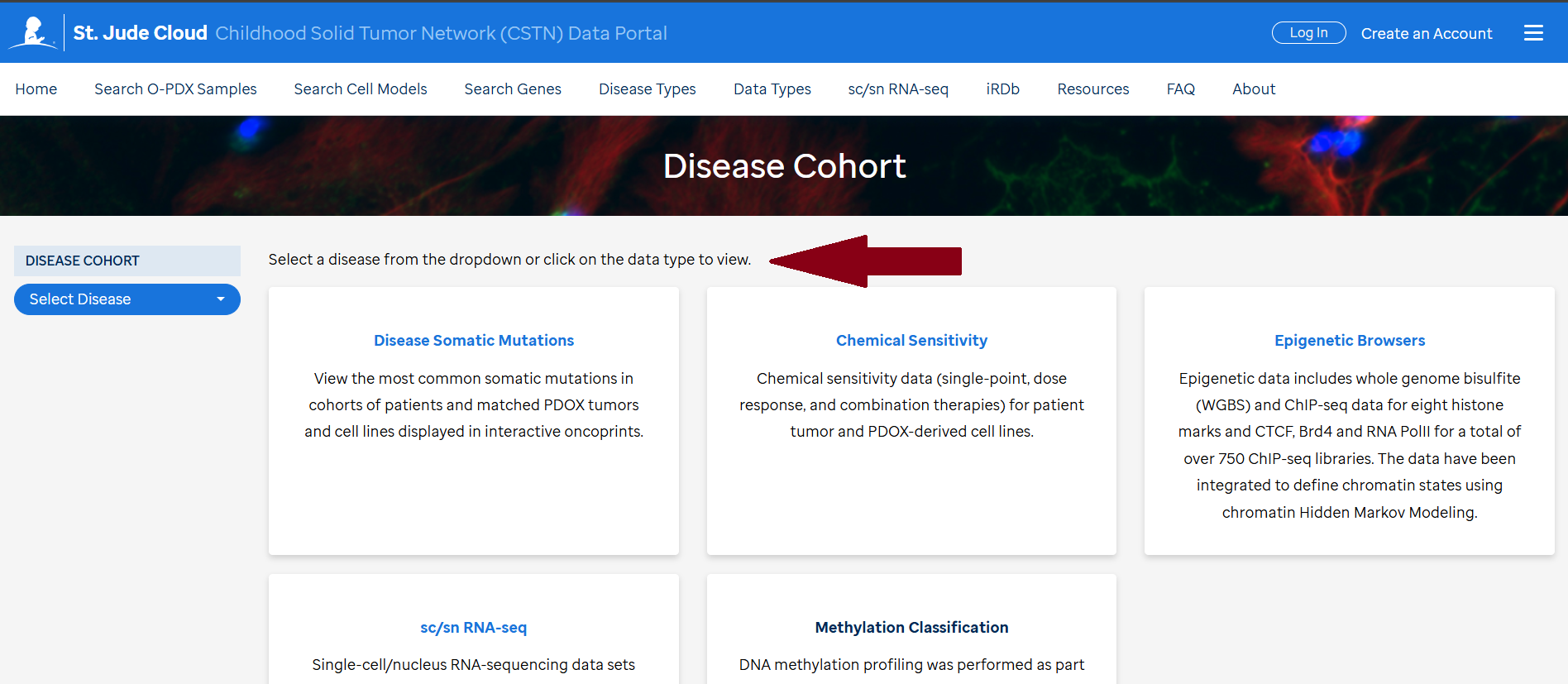
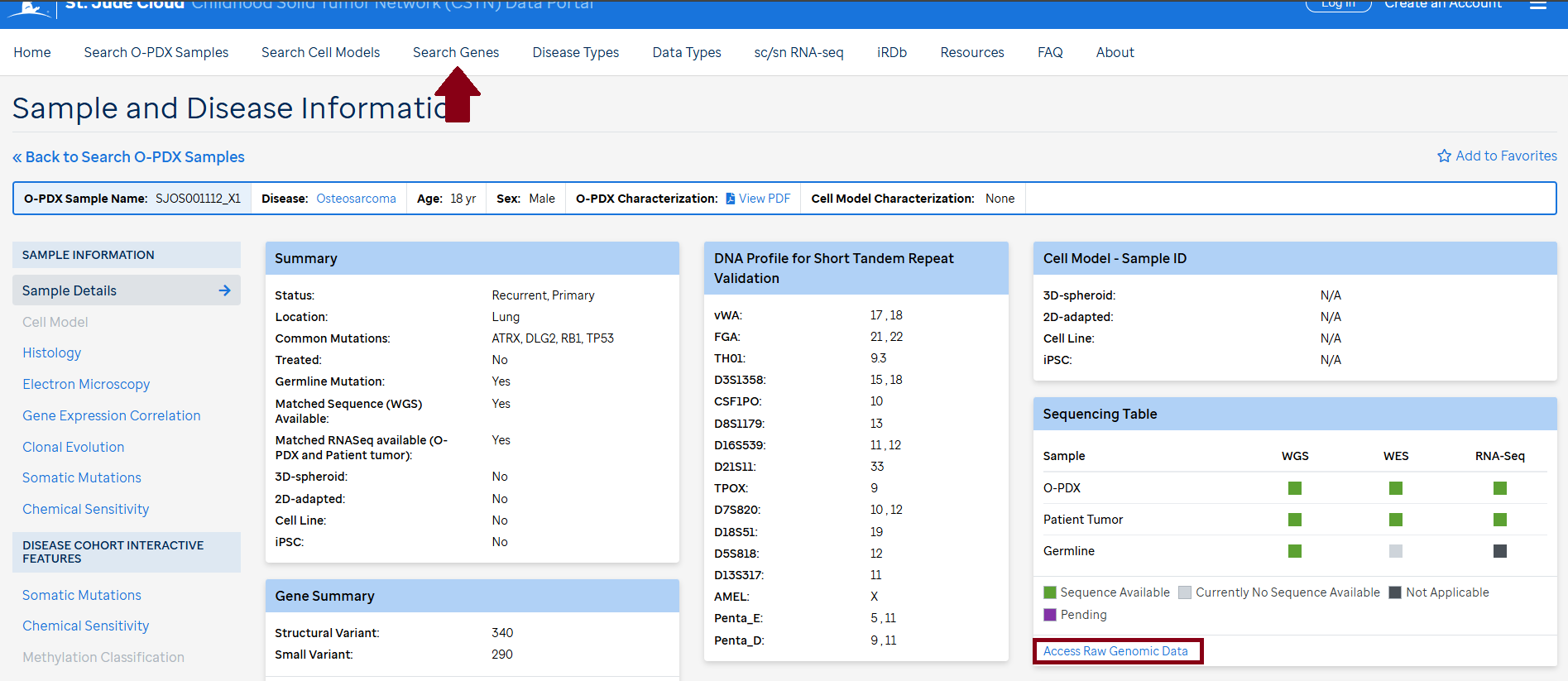
Bam files for WGS, WES and RNASeq for the O-PDX samples, corresponding primary tumors and germline (if available) can be found on SJCloud under the CSTN tag. There is a quick link (Access Raw Genomic Data) in each sample on the CSTN data portal. To access the bam files the requester needs to follow the SJCLoud DAA process.
Access to curated data is available in the CSTN data portal under "Search Genes" and each individual sample "Access Raw Genomic Data".